I just realized that it's been a long time since I blogged activity 7, also meaning that it's been a long time since I started doing activity 8. Sorry ma'am for the delay. Don't worry, I'm excited for what's in store in activity 9.
Activity 8 is about a new topic -- morphological operations. Morphology, according to Merriam-Webster dictionary (http://www.merriam-webster.com/dictionary/morphology) is the study of structure and form. Images always contain these elements of structure and form, and in this activity, we focus on these. Part of processing images is changing the form of an object, and this can be done using morphological operations. Classical morphological operations are used on binary images (according to our manual prepared by ma'am Soriano), which are usually depicted as black and white images. Here comes in the importance of image segmentation, which we have dealt with in our previous activity.
The discussion in our manual on morphological operations contains set theory and explains the morphological operations in a mathematical way. Although this is the formal way of describing these operations, I would want to explain these in a different manner for easier understanding, which is basically adapted from these links
http://homepages.inf.ed.ac.uk/rbf/HIPR2/erode.htm
http://homepages.inf.ed.ac.uk/rbf/HIPR2/dilate.htm
In using morphological operations, we want to operate on images which contain background (usually black) and foreground (usually white) pixels. We also need a structuring element, which is also a binary matrix, that determines the extent of the effect of the operation. The elements that will matter in the structuring element are the foreground pixels. For the structuring element, we assign one of its elements (or even an element outside the matrix) as an origin.
We now discuss one morphological operation -- erosion. To perform this, we coincide the origin of the structuring element with a foreground pixel of the image, say \(x\). If all of the foreground pixels in the structuring element coincide with foreground pixels in the image, then \(x\) is retained as is. If not, then \(x\) is turned to a background pixel. This is done for all of the foreground pixels of the image.
Opposite to erosion is dilation. For this operation, we coincide the origin of the structuring element with a background pixel in the image, say \(x\). If a foreground pixel in the structuring element coincides with a foreground pixel in the image, then \(x\) is turned into a foreground pixel. If there are none, \(x\) is retained as a background pixel. This is performed for all background pixels in the image.
Now that we know 2 basic morphological operations, we go to the activity proper. The first part of the activity was to test our knowledge on erosion and dilation. We were tasked to illustrate by hand drawing the erosion and dilation of 4 different shapes using 5 different structure elements. The shapes are
A. 5x5 square,
B. triangle with base = 4, height = 3 boxes,
C. hollow 10 x 10 square, 2 boxes thick, and
D. plus sign, 1 box thick, and 5 boxes along each line.
The structuring elements are
1. 2x2 ones,
2. 2x1 ones,
3. 1x2 ones,
4. cross, 3 pixels long, 1 pixel thick, and
5. a diagonal line (lower left to upper right), 2 boxes long.
These shapes and structuring elements are illustrated below
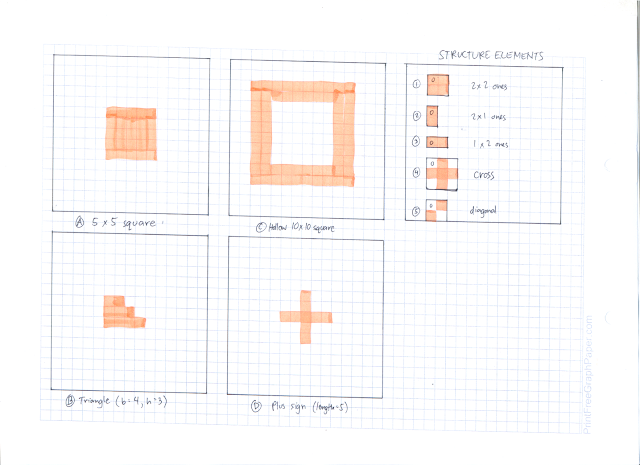 |
| Shapes and structuring elements used |
Now we perform erosion on the different shapes. Note that on the images, the letter denotes the shape being operated on and the number denotes the structuring element used.
Erosion of the shapes
On the other hand, the following illustrations demonstrate dilation operation.
Dilation of the shapes
To verify whether our understanding of the erosion and dilation operations are correct, we use Scilab to perform these operations. These operations are available in the Image Processing Design toolbox of Scilab by calling ErodeImage and DilateImage functions. The resulting images are shown below. Again note that the numbers denote the structuring element used as labeled above.
 |
| Erosion and dilation using Scilab |
We observe that the results are the same when we use structuring elements 1 - 3, but differ when 4 and 5 are used. This is due to a difference in the set origin of the structuring element. The origin of the structuring element affects the resulting morphology after operation. In the case of the cross in Scilab, it seems that the origin was located at the center of the cross. For the diagonal, the origin was located a the lower left pixel of the diagonal. These can be deduced by observing the resulting images using structuring elements 4 and 5.
Now we are to apply morphological operations in processing images. We have below an image of simulated cells (punched holes) and what we want to do is obtain an estimate size of these cells. To start, we segment this image to determine the cells from the background.
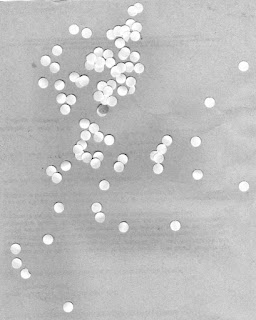 |
| Simulated cells |
The histogram of the gray values in the image is shown below so that we can determine a threshold value to use. The values within the band correspond to the background of the image. We observe from the image that the cells are lighter than the background, so we deduce that the values greater than the band correspond to the cells.
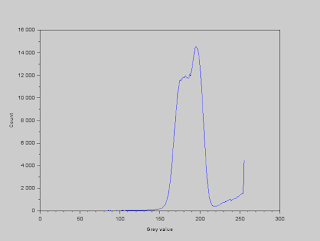 |
| Histogram of gray values |
We set the threshold gray value to 210, and segment the image by using the function SegmentByThreshold. All values greater than this threshold are set to white and all values less than this threshold are set to black. The resulting image is shown below.
 |
| Thresholded image |
Here we see unwanted dots scattered in the image which are not cells. These are unwanted features in our segmented image. To remove these, we need to use morphological operations. We introduce another morphological operation -- opening. The opening operation is simply a combination of the operations we studied above. This operation involves eroding first the image and then dilating it. If we, for example, have a structuring element that is larger than a foreground region in the image, then erode the 2 images, the region completely turns into a background region. If we then perform dilation, the original foreground region would not be recovered. Effectively, with the opening operation, we can open gaps between the foreground pixels or we can see it as eliminating foreground pixels (http://homepages.inf.ed.ac.uk/rbf/HIPR2/open.htm). In our case, we use the OpenImage function using a circular structuring element with a radius of 4 pixels. The resulting image is shown below.
 |
| Thresholded and cleaned image |
We have now isolated the features we want to work with. The next thing we have to do is measure the sizes of each cell. To start, we identify each cell in our image, or blobs in general, and we do this by using the function SearchBlobs. What this does is label each blob with a number, and we can use that number to isolate that blob. At this point, when we use this function, we note that some of the cells are overlapping, which will effectively constitute a single blob. For the moment, we ignore this fact and calculate the area of each blob. The histogram of the blobs' areas (in pixels) are shown below.
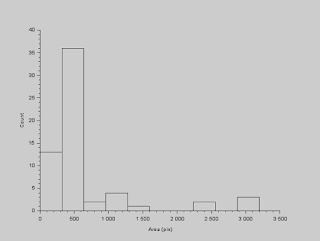 |
| Histogram of cell areas |
To deal with this wide range of values for the areas, we use statistics! What we can do is determine the outliers in this data set and remove them. We can do this by using interquartile range (IQR). Outliers in a data set are those values (say \(x\)) which are \(x < Q_1 - 1.5IQR \) or \(x > Q_3 + 1.5IQR \), where \(Q_1\) is the 1st quartile, \(Q_3\) is the 3rd quartile, and \(IQR = Q_3 - Q_1\). After removing the outliers, the histogram results to the plot below.
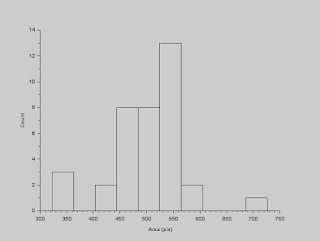 |
| Filtered histogram of cell areas |
We can now calculate for an estimate of the cells' sizes. The mean is 501.08 and the standard deviation is 71.49.
Now that we have an estimate of the cells' sizes, we can now go on to identify cancer cells among the normal cells. The image shown below contains normal cells and cancer cells. Initial steps in processing this image are the same for the previous image. We segment this image and clean it to remove the unwanted features.
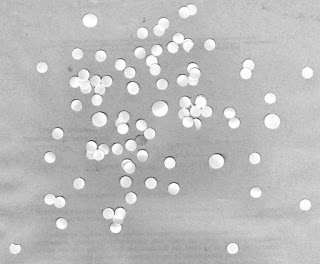 |
| Simulated normal and cancer cells |
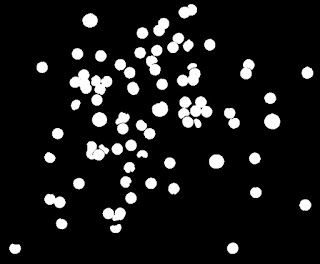 |
| Binarized image of the cells |
We then identify the blobs in the image. The image below shows the identified blobs, where each blob denotes a different shade from the others.
 |
| Image with identified blobs |
Now, we use our estimate earlier to filter out the normal cells. We do this using the FilterBySize function. We set our threshold to a size of 501.08 + 71.49 = 572.57. When the size is lesser than this value, the blob is set to a background, and when the size is greater than this value, the blob is retained. We observe now that we are left with overlapping normal cells and abnormally large cells, which we deduce to be the cancer cells.
 |
| Image with filtered blobs by size |
Now, what we are left to do is to remove these overlapping normal cells, and to do this, we use the magic of the opening operation. We can create a structuring element that can remove these but not the cancer cells. In this case, a circular structuring element with a radius of 13 pixels is appropriate. The cancer cells are now successfully identified as shown below (binarized image).
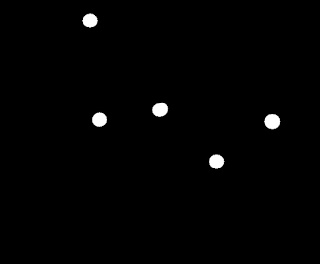 |
| Cancer cells |
I would give myself a grade of 10 because I have done all of the stated tasks for this activity.



















































